Caterva2: On-demand access to Blosc2/HDF5 data repositories#
What is it?#
Caterva2 is a service for sharing Blosc2 and HDF5 datasets with authenticated users, work groups, or the public. It provides multiple interfaces: web GUI, REST API, Python API, and command-line client.
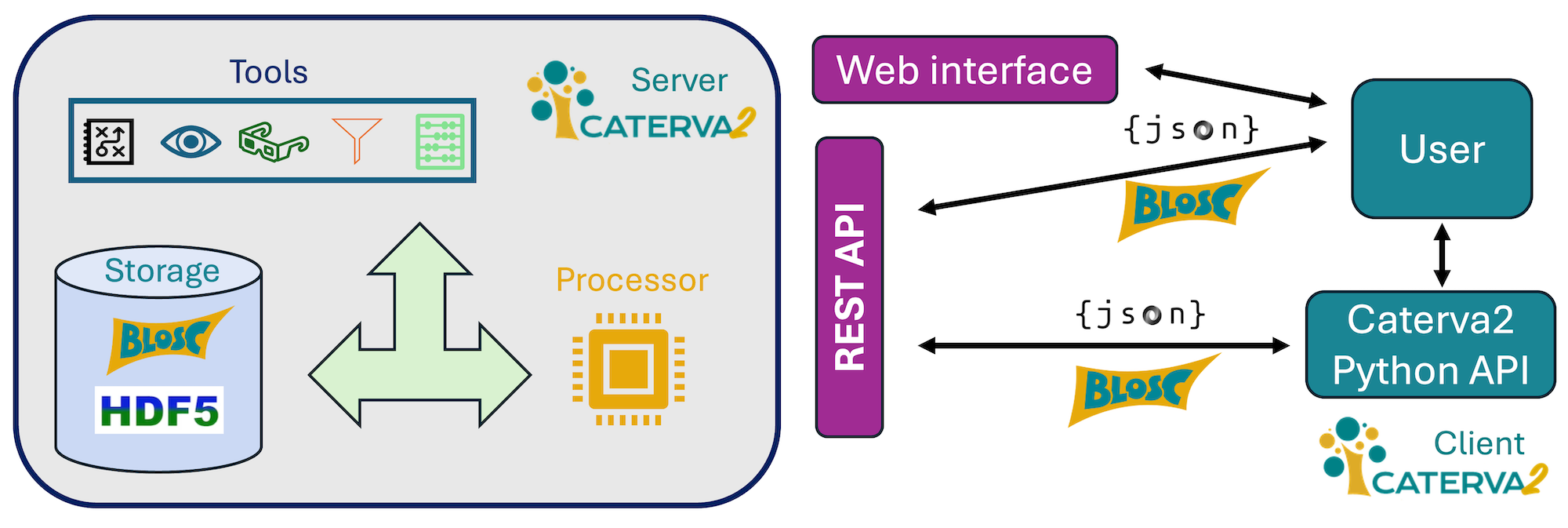
Use it remotely or locally to access datasets in a directory hierarchy or share them across your network.
The Python API is recommended for building custom clients, while the web GUI offers a user-friendly interface for browsing datasets.
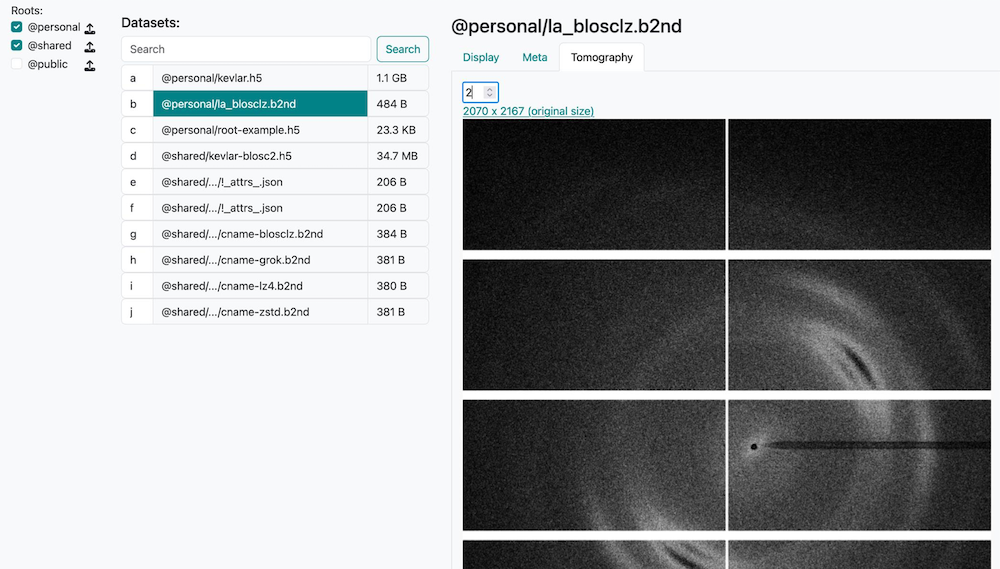
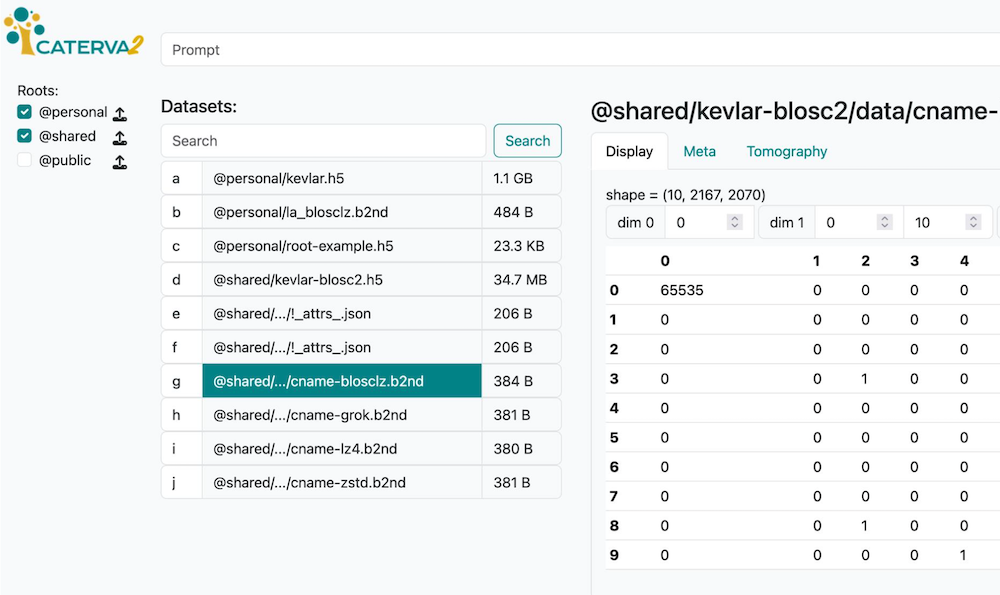
Caterva2 Clients#
The Caterva2 package provides a lightweight library for building custom clients. Choose the interface that best fits your needs:
Web GUI - Browser-based interface
Python API - Programmatic access
import caterva2 as cat2 client = cat2.Client("https://cat2.cloud/demo") print(client.get("@public/examples/numbers_color.b2nd")[2])
Command-line client - Terminal interface
cat2-client --server https://cat2.cloud/demo info @public/examples/numbers_color.b2nd
REST API - HTTP interface (use with Postman, curl, etc.) See the live API docs at cat2.cloud/demo/docs.
All interfaces support authentication for accessing private datasets (see “User authentication” below).
Installation#
For Users#
Client only (Python API and CLI tools):
pip install caterva2[clients]
Test the installation (includes client, server, and test suite):
pip install caterva2[tests]
python -m caterva2.tests
CATERVA2_SECRET=c2sikrit python -m caterva2.tests # with authentication
For Developers#
Install from source (includes server, clients, and test suite):
git clone https://github.com/ironArray/Caterva2
cd Caterva2
pip install -e .[tests]
python -m pytest
CATERVA2_SECRET=c2sikrit python -m pytest # with authentication
Available Extras#
Append [extra1,extra2,...] to any install command:
clients- CLI and terminal toolsserver- Server servicetests- Test suite (includes server and clients)blosc2-plugins- JPEG 2000 support via blosc2-grok
Note: Test runs create a _caterva2_tests directory with state files for inspection.
Quick start#
See Caterva2 documentation for detailed tutorials.
Setup:
Install with server and client support:
pip install caterva2[server,clients]
Copy configuration files:
caterva2.sample.toml→caterva2.toml(client config)caterva2-server.sample.toml→caterva2-server.toml(server config)
Place in current directory,
~/, or/etc/. Use--confto specify alternate locations.Start the server:
CATERVA2_SECRET=c2sikrit cat2-server &
CATERVA2_SECRETis required for user authentication (see below).
Server directory structure:
tree _caterva2
_caterva2
└── state
├── db.json # metadata
├── db.sqlite # metadata
├── media # temporary files for web GUI
├── personal # user-specific datasets
├── public # publicly shared datasets
└── shared # group-shared datasets
Populate with example datasets:
cp -r root-example/ _caterva2/state/public/
Browse them at http://localhost:8000/?roots=@public
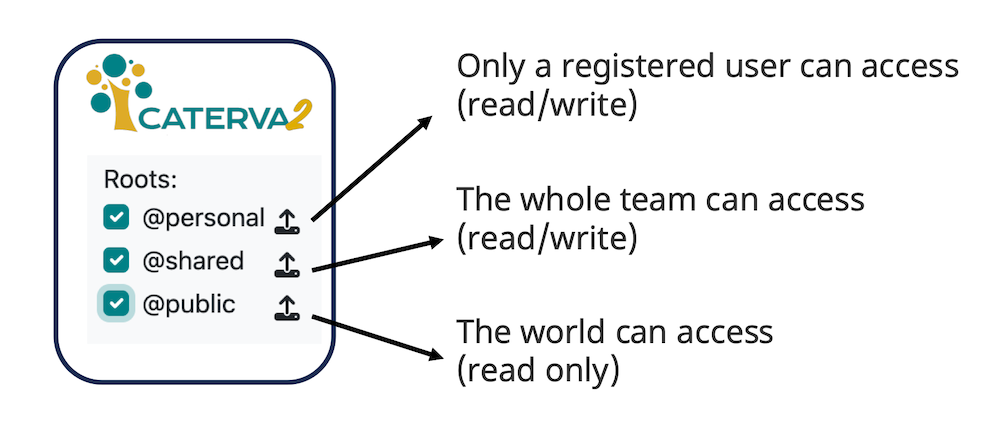
User authentication#
Enable authentication by setting CATERVA2_SECRET when starting the server. This enables user management but does not verify email addresses or support password recovery.
Create a user:
cat2-admin adduser user@example.com foobar11
Authenticate clients:
Web GUI - Login prompt on access
Python API - Pass credentials to client:
client = cat2.Client("http://localhost:8000", ("user@example.com", "foobar11"))
CLI - Use
--userand--passoptions
Command-line client#
List available roots:
cat2-client --user user@example.com --pass foobar11 roots
Show output
@public
@personal
@shared
List datasets:
cat2-client list @public
Show output
examples/README.md
examples/Wutujing-River.jpg
examples/cat2cloud-brochure.pdf
examples/cube-1k-1k-1k.b2nd
examples/cubeA.b2nd
examples/cubeB.b2nd
examples/dir1/ds-2d.b2nd
examples/dir1/ds-3d.b2nd
examples/dir2/ds-4d.b2nd
examples/ds-1d-b.b2nd
examples/ds-1d-fields.b2nd
examples/ds-1d.b2nd
examples/ds-2d-fields.b2nd
examples/ds-hello.b2frame
examples/ds-sc-attr.b2nd
examples/gaia-ly.b2nd
examples/hdf5root-example.h5
examples/ironpill_nb.ipynb
examples/kevlar-tomo.b2nd
examples/lazyarray-large.png
examples/lung-jpeg2000_10x.b2nd
examples/numbers_color.b2nd
examples/numbers_gray.b2nd
examples/sa-1M.b2nd
examples/slice-time.ipynb
examples/tomo-guess-test.b2nd
large/gaia-3d.b2nd
large/slice-gaia-3d.ipynb
Browse directory tree:
cat2-client tree @public
Show output
├── examples
│ ├── README.md
│ ├── Wutujing-River.jpg
│ ├── cat2cloud-brochure.pdf
│ ├── cube-1k-1k-1k.b2nd
│ ├── cubeA.b2nd
│ ├── cubeB.b2nd
│ ├── dir1
│ │ ├── ds-2d.b2nd
│ │ └── ds-3d.b2nd
│ ├── dir2
│ │ └── ds-4d.b2nd
│ ├── ds-1d-b.b2nd
│ ├── ds-1d-fields.b2nd
│ ├── ds-1d.b2nd
│ ├── ds-2d-fields.b2nd
│ ├── ds-hello.b2frame
│ ├── ds-sc-attr.b2nd
│ ├── gaia-ly.b2nd
│ ├── hdf5root-example.h5
│ ├── ironpill_nb.ipynb
│ ├── kevlar-tomo.b2nd
│ ├── lazyarray-large.png
│ ├── lung-jpeg2000_10x.b2nd
│ ├── numbers_color.b2nd
│ ├── numbers_gray.b2nd
│ ├── sa-1M.b2nd
│ ├── slice-time.ipynb
│ └── tomo-guess-test.b2nd
└── large
├── gaia-3d.b2nd
└── slice-gaia-3d.ipynb
Get dataset info:
cat2-client info @public/examples/ds-1d.b2nd
Show output
Getting info for @public/examples/ds-1d.b2nd
shape : [1000]
chunks: [100]
blocks: [10]
dtype : int64
nbytes: 7.81 KiB
cbytes: 4.90 KiB
ratio : 1.59x
mtime : 2026-01-15T17:04:50.823466Z
cparams:
codec : ZSTD (5)
clevel : 1
filters: [SHUFFLE]
For more commands: cat2-client --help
Documentation#
For tutorials, API references, and examples, visit the Caterva2 documentation.
